| Ohio State University Medical Center |
| Advanced Screening for Active Protocols |
"Iron Speed Designer easily accommodated frequent user requirement
changes, ranging from web page layout to the underlying data model."
- Jing Ding, Senior Systems Consultant of Information Warehousing at OSUMC
|
|
|
Advanced Screening for Active Protocols |
Ohio State University Medical Center
Columbus, Ohio USA
|
The Advanced Screening for Active Protocols (ASAP) application was created for the Ohio
State University Medical Center (OSUMC). OSUMC is located in Columbus, Ohio, and is one
of the largest and most diverse academic centers in the country. It is the only academic
medical center in central Ohio.
ASAP is used within OSUMC by medical screeners and clinical trial coordinators who look for
patients to enroll in clinical studies.
This new application searches patients' electronic medical records (EMRs) using specific
screening criteria to see if they are eligible for the clinical studies. The first step
is for a screener or coordinator to register the study in ASAP via a typical Add Record
page.
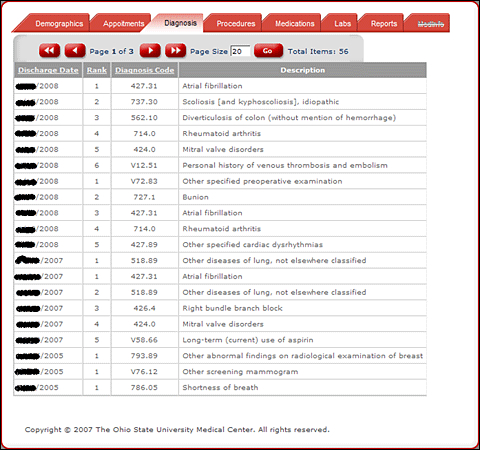
Patient EMR.
Once the study is registered, the user enters its eligibility criteria; for example, male,
age between 18 and 70, diagnosed with diabetes, etc. A study may have more than ten criteria.
Each criterion is a query template pointing to one of the EMR tables. The user selects a
combination of the templates and fills in parameters via a customized Edit Table page.

Setup study eligibility criteria.
The criteria are run against patient EMRs on a daily basis for the duration of the study.
Qualified patients are summarized in a Telerik RadGrid control, which has rows representing
patients and columns representing criteria. The grid cells link to various Show Record and
Show Table pages that display relevant portions of patient EMRs. From here, the user identifies
candidate participants.
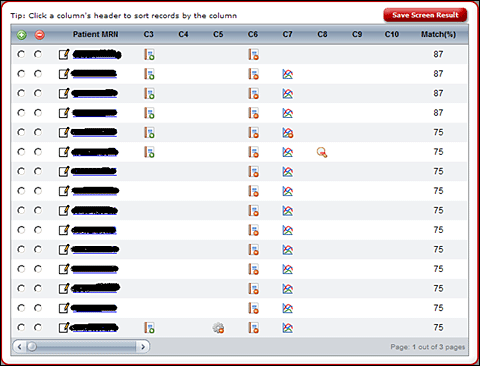
Screen result: potentially eligible patients.
|
Application size and scope |
The Advanced Screening for Active Protocols (ASAP) accesses seven EMR-related databases
and one ASAP-specific database, using Oracle. The ASAP-specific database contains 18
tables and 11 views. In addition, ASAP accesses the other seven databases through 14
materialized views. The largest database table has approximately 125 million rows, which
equates to 1 million rows in its corresponding materialized view. The application has 36
Web pages, including ASCX controls.
In its initial phase, ASAP serves five ongoing medical studies. In the future, it may
serve hundreds or even thousands of studies.
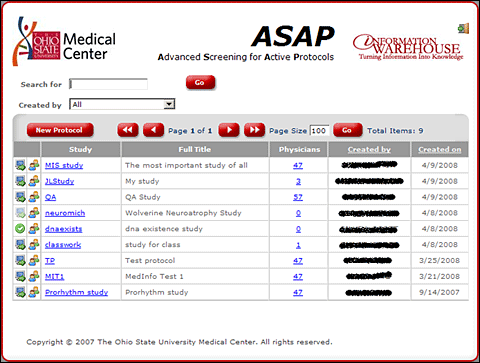
List of on-going studies.
|
The project |
It took one person two months to develop the first working prototype of this project. After
that, there were multiple changes based on user feedback. Each development iteration took
about one week.
My time was spent 95% on page development, 20% of which was in Iron Speed Designer which
I used to build the framework of this application. I spent the rest of the time customizing
my application in Microsoft Visual Studio.
|
Code extensions and customizations |
The ASAP application is extensively customized. I wrote approximately 5,000 lines of code
in the following areas, among others:
 | Large list selectors |
 | Email |
 | Conditional display |
 | Custom queries at runtime |
 | Ajax functions |
 | ASCX controls
|
The code customization took 80 percent of the development time. In addition to in-house
code customization, I incorporated several Telerik controls including: RadGrid, RadWindow,
RadCalendar, RadTextBox and RadComboBox.

Setup study eligibility criteria.
|
Page layout customizations |
I made extensive page customizations, including:
 | Rearranging control layout |
 | Adding additional record, table, ASP or ASCX controls |
 | Printable page |
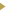 | Integrating third-party controls
|
I used the standard Fuji design theme in Iron Speed Designer.
|
Iron Speed Designer impact |
|
Iron Speed Designer easily accommodated frequent user requirement
changes, ranging from web page layout to the underlying data model.
|
Next steps |
|
We have plans to expand the ASAP application in the future. As the application is
rolled out to additional users, we will add more query templates. There is also a
possibility of adding additional sites.
|
About the developer |
|
Jing Ding has a PhD in Computer Engineering, Bioinformatics and Computational Biology,
and an M.S. in Toxicology from Iowa State University. He received his B.S. in Biophysics
from Fundan University in Shanghai, China. He is a self-taught programmer who "played"
with assembly, C and C++ in the 1990s. He took a break from programming from 1997 to 2000.
When he picked it up again in 2001, he worked with Java. Jing began working with C#.NET in 2006.
|
|
|
|
|
|
|
